Projects
Research Opportunities
I am always interested in having enthusiastic and passionate students in my laboratory if there are openings. Students in my laboratory will be exposed to a variety of techniques from many disciplines including; biochemistry, molecular biology, enzymology, microbiology, plant biology/pathology, evolutionary biology, structural biology among others. Email me at aohsbi@rit.edu to inquire about research opportunities.


Amino Acid Metabolism
My laboratory is primarily interested in amino acid metabolism. More specifically, we are interested in the aminotransferase class of enzyme. Aminotransferases or transaminases (EC 2.6.1.x) are ubiquitous enzymes that are involved in amino acid biosynthesis, vitamin metabolism, carbon and nitrogen assimilation, secondary metabolism etc. They catalyze reversible reactions by transferring an amino group from a donor to an acceptor. The amino donor is usually an amino acid and the amino acceptor is usually a 2-oxo-acid. In the model organism Arabidopsis thaliana, there are 44 annotated aminotransferases many of which are uncharacterized. Using biochemical and bio-informatical approaches, my lab is interested in elucidating the function of the remaining aminotransferase enzymes that are deemed putative from organisms such as; plants, bacteria and algae.


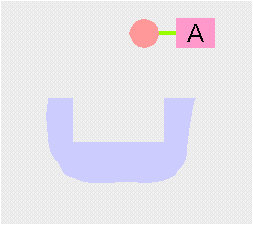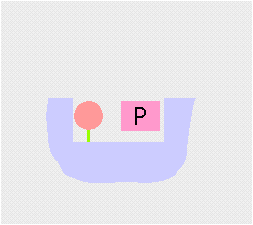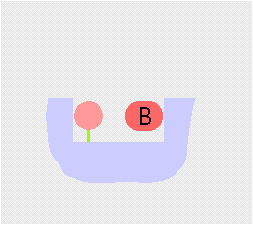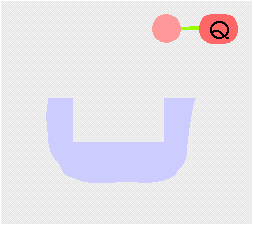
Transamination interconverts pairs of amino acids and keto acids. During transamination, the amino group of an amino acid is transferred to a keto acid, this produces a new keto acid while from the original keto acid, a new amino acid is formed. The enzyme employs a ping-pong (double displacement)(see cartoon above) mechanism facilitated by the co-factor pyridoxal phosphate (PLP) which is bound to a conserved lysine residue in the active site of the enzyme.
Structural Analysis of Enzymes Involved in Amino Acid Metabolism/ Characterization Novel Targets for Antibiotic Development
With Dr. Renwick C.J. Dobson - University of Canterbury, New Zealand
Some of the enzymes involved in the metabolism of amino acids are putative or validated antibiotic targets based on the fact these amino acids are essential for bacterial growth and the anabolic pathways are absent in animals particularly humans. The Hudson lab is currently collaborating with the lab of Dr. Renwick Dobson from the University of Canterbury to elucidate the 3-dimensional structure of enzymes involved in amino acid metabolism.
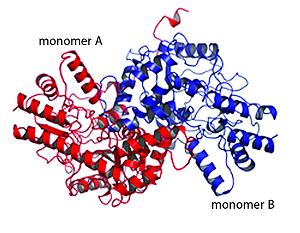
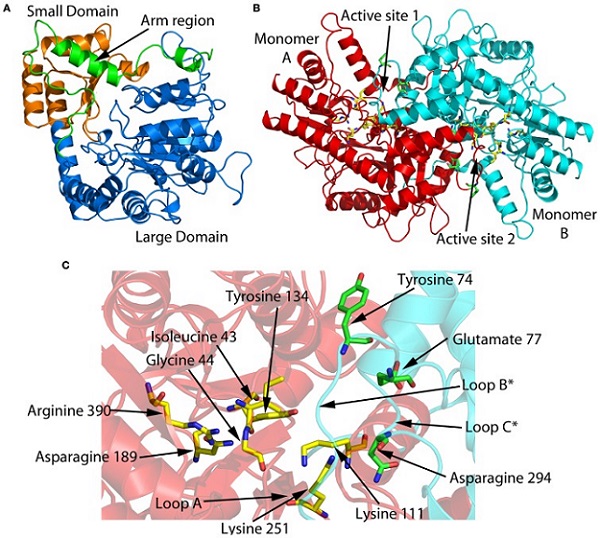
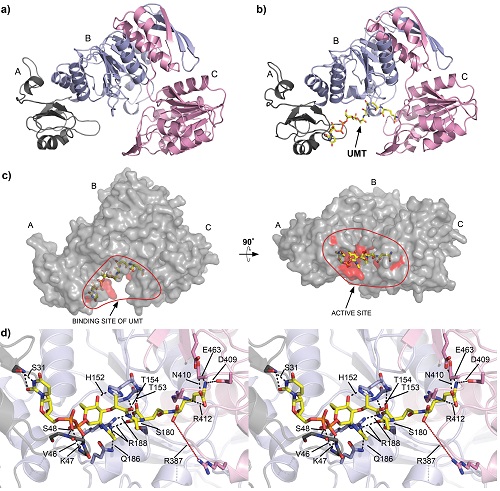
Identification of Bacterial Endophytes/Epiphytes
A collaborative project with Dr. Michael Savka (GSoLS) to identify and assess the role(s) of endophytic and epiphytic bacteria from plants such as; sugarcane, grape etc is under way.
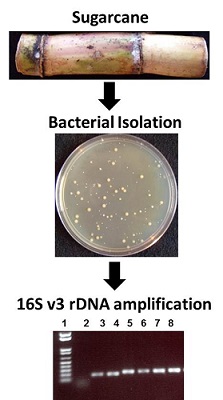
Isolation and identification of bacterial endophytes from sugarcane using 16S V3 rDNA analysis.
Recently, the genomes of 19 plant associated bacteria fromw sugarcane, yam and willow were sequenced and annotated in collaboration with Dr. Han Ming Gan (Monash University, Malaysia).
Acknowledgements
I would like to thank the National Institutes of Health (NIH), the National Science Foundation (NSF), The RIT College of Science (COS), The Thomas H. Gosnell School of Life Sciences (GSoLS), The RIT Office of the Vice President for Research (OVPR), Bayer Corporation, Sweetwater Energy, Natcore Technology and Iridium Biosciences for financial support. I would also like to thank all of my internal and external collaborators, undergraduate, graduate and post-doctoral colleagues for their wonderful contributions to the various projects in my lab.