Andre Hudson
Dean, College of Science
Andre Hudson
Dean, College of Science
Education
BS, Virginia Union University; Ph.D., Rutgers University
Bio
Dr. Hudson is trained as a biochemist and the major themes of his research are vested in biochemistry and microbiology. More specifically, in the areas of amino acid metabolism, structural analyses of enzymes involved in amino acid and bacterial peptidoglycan metabolism, and the isolation, identification and genomic characterization of plant-associated bacteria. Dr. Hudson has secured approximately $1.3 million in federally funded grants and contracts as PI and or CoPI from the NIH, NSF, Bayer Corporation, Sweetwater Energy and Natcore Technology. Dr. Hudson has published 43 peer-reviewed articles, and presented more than 28 conference presentations in addition to 29 invited talks.
Dr. Hudson is a highly respected and well liked teacher. His teaching contributions are substantial especially during the conversion to semesters when he rewrote all the courses he teaches. Dr. Hudson has mentored and engaged many students in research and has published in peer-reviewed journals with a number of them. Many of his students have gone to pursue further research at prestigious institutions.
Since joining RIT, Dr. Hudson has served on numerous School, College, and Institute Committees, including the GSoLS Curriculum Committee, the College of Science Faculty Evaluation and Development Committee which he has chaired since 2014, and the RIT Isaac L. Jordan Faculty Pluralism Award Selection Committee which he has chaired since 2013.
Dr. Hudson joined the RIT faculty in 2008 following a post-doctoral fellowship at Rutgers University. He earned his B.S. (2000) in Biology from Virginia Union University, Richmond, VA., and his Ph.D. (2006) in Plant Biochemistry from Rutgers University.
Select Scholarship
Currently Teaching
In the News
-
January 19, 2026

Four advances reshaping the fight against antibiotic resistance
An essay by André Hudson, dean of the College of Science, published by The Washington Post. (This content may require a subscription to view.)
-
October 29, 2025

Simons Empire Faculty Fellowship to fund four College of Science positions
RIT’s College of Science will receive funding from the Simons Empire Faculty Fellowship program for four new tenure-track faculty members.
-
July 15, 2025

Lousto honored with Argentine award for outstanding scientific contributions
Carlos Lousto, Distinguished Professor in the School of Mathematics and Statistics, is a recipient of the 2024 RAICES Award, granted by the Secretariat of Innovation, Science, and Technology (SICyT) of the Argentine government.
-
April 10, 2025
2025 Celebration of Teaching and Scholarship
-
August 29, 2023
Hudson named senior editor for ASM
-
June 20, 2022
RIT hosts 2022 GENIUS Olympiad
-
December 14, 2021
Team publishes paper on antibiotic-producing bacteria
Featured Work
Accessible First-Year Research Opportunities
McKenzie Watts (biotechnology)
McKenzie Watts, a first-year student, says the best part about RIT is that research is accessible. Today, she's working in a research lab with Dr. André Hudson.
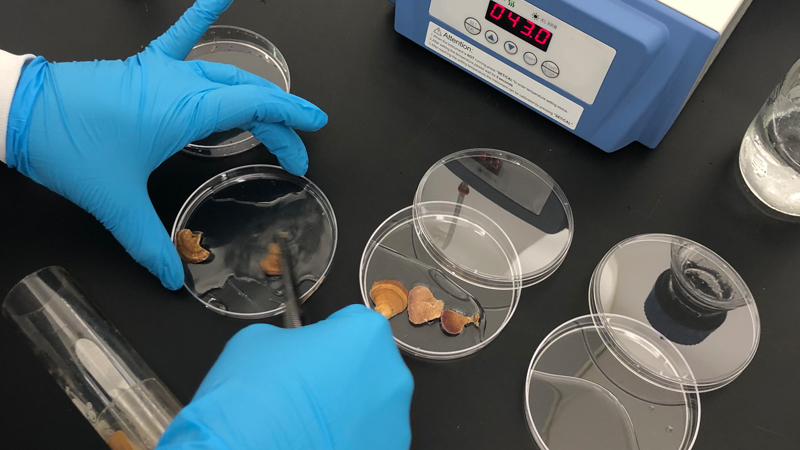
Genomics Students Co-Author Published Paper in American Society for Microbiology
Andre Hudson
Students in the RIT undergraduate genomics course designed their semester long collaborative project on wild mushrooms and were given the opportunity to co-write a published paper.