Cui Research Group

Cui Research Group
- RIT/
- College of Science/
- Research/
- Cui Research Group
Contact
Feng Cui, Ph.D.
Associate Professor
College of Science
Thomas H. Gosnell School of Life Sciences
Email
Are you a passionate, forward-thinking individual with a knack for computer science and an insatiable curiosity for the world of biology and medicine? Join the Cui Research Group in the exciting field of Artificial Intelligence (AI) and Biology.
About Dr. Feng Cui

Feng Cui, Ph.D. is an Associate Professor in the RIT Thomas H. Gosnell School of Life Sciences. He earned a Ph.D. in Bioinformatics and Computational Biology at Iowa State University and subsequently spent several years as a postdoctoral fellow at National Cancer Institute (NCI). His research is mainly focused on the nucleosome and its associated binding proteins. Dr. Cui serves as the Graduate Director of the Bioinformatics MS Program and holds the position of Faculty Senator (Alternate). He enjoys working with students at the interface of Biology and Computer Science. Dr. Cui is also an affiliated faculty member of the Golisano College for Computing and Information Sciences.
Seeking Undergraduate and Graduate Students
The Cui Research Group is seeking BS, MS and Ph.D. students who demonstrate a passion for learning and possess coding abilities. As we continue to push the boundaries of knowledge in our field, we invite you to join us and make a valuable contribution to our cutting-edge research. Do you have the following requirements and want to work in a dynamic and collaborative environment?
- Demonstrated coding skills.
- Interest in gaining valuable experience.
- Enthusiasm for learning and research.
- Commitment to the project.
If you meet these requirements we encourage you to apply for a position in our lab.
Current Projects
1. Exploring the diversity of nucleosomal DNA
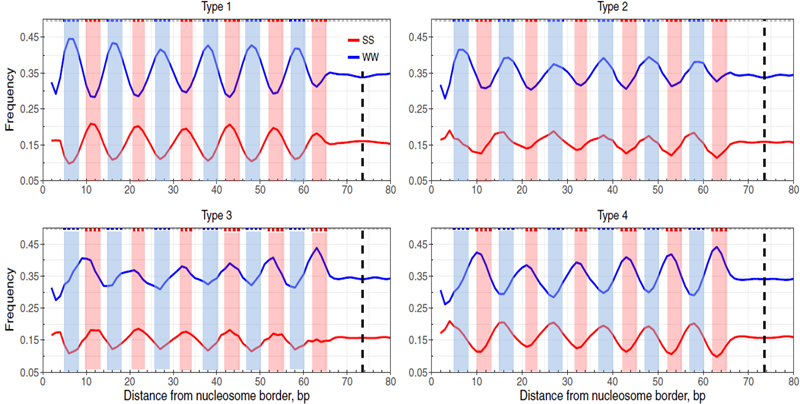
The basic repeating unit of chromatin is the nucleosome core particle, which consists of a histone octamer, around which 147 bp DNA are wrapped about 1.7 turns. Structurally, the 147-bp DNA fragment can be divided to two halves by the dyad at position 74. Each half is consisted of six minor-groove bending sites (GBS) (at superhelical locations (SHL) -6.5 to -1.5) where DNA is bent into minor grooves and six major-GBS (SHL -6 to -1) where DNA is bent into major grooves.
Four types of DNA patterns have been observed in nucleosomal DNA. Type 1 is the pattern described by Travers and colleagues, in which WW dimers have peaks at minor-GBS (blue shading), and SS dimers have peaks at major-GBS (red shading). Type 2 is the pattern in which both WW dimers and SS dimers have peaks at minor-GBS. Type 3 is the pattern in which both WW dimers and SS dimers have peaks at major-GBS. Type 4 is opposite to Type 1, in which WW dimers have peaks at major-GBS, and SS dimers have peaks at minor-GBS. Here, we refer to Type 1 as the well-known ‘WW/SS pattern’ and Type 4 as the ‘anti-WW/SS pattern’.
Our work aims to understand the structural basis and functional significance of the nucleosomes with Type 4 pattern (i.e., anti-WW/SS nucleosomes). We have found that anti-WW/SS nucleosomes account for 13-31% of all nucleosomes in different species and are widespread across the genomes. They are enriched in genic regions in mammals and correlated with transcriptional levels. Currently, we are trying to understand whether they are also enriched around transcription factor binding sites and how their structures are stabilized.
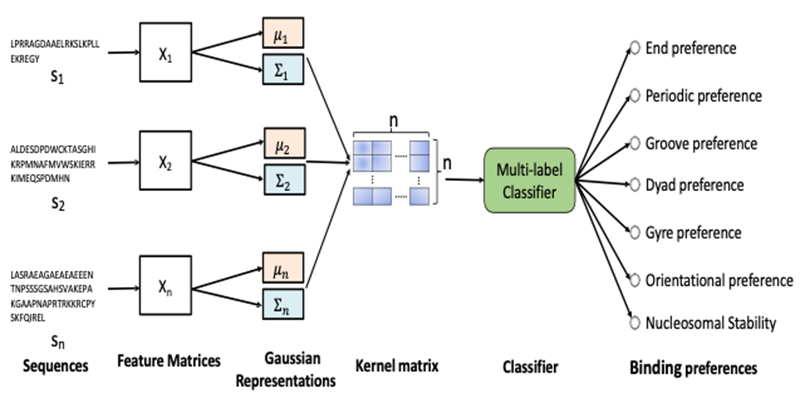
2. Predicting nucleosome-binding proteins by machine learning
A small group of TFs known as pioneer TFs are able to bind nucleosomal DNA and play an important role in cell development. Based on the Systematic Evolution of Ligands by Exponential Enrichment (SELEX) technique, recent studies have identified several TF-nucleosome interaction modes including end binding, oriented binding, periodic binding, dyad binding, groove binding, and gyre spanning. However, there are substantial experimental challenges in measuring nucleosome binding modes for thousands of TFs in different species. In this study, we aimed to test a hypothesis that the binding modes of a TF to a nucleosome can be predicted by their amino acid sequences using machine learning methods. To achieve this aim, we have developed a machine learning model, ProtGauss, to predict the nucleosome binding modes based on an unsupervised data-driven Gaussian representation for protein sequences. Currently, we are developing novel deep learning frameworks to predict nucleosome-binding proteins in a specie-specific manner.
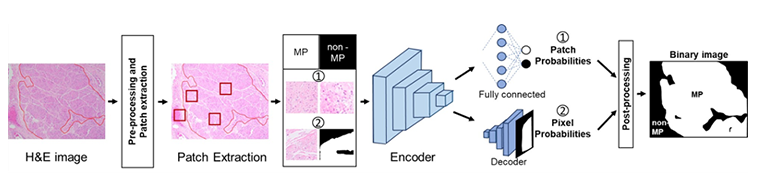
3. AI in medicine and systems biology
The biological system is a complex network of heterogeneous molecular entities such as genes, proteins, and other biomolecules linked together by their interactions. Although enormous technological advancements have been made in the past four decades, experimental determination of these interactions has been a great challenge. With the ever-increasing computational resources and novel methodologies capable of handling large datasets, we developed a suite of algorithms based on deep learning to represent biological entities and model their interactions. These methods accurately predicted the interactions between genes, between proteins, and between drugs and targets. Moreover, we developed novel deep learning frameworks to predict bladder cancer types, smooth muscle fibers, as well as biomarkers associated with distant metastasis of various carcinoma. Currently, we are developing novel AI models to predict novel sex-associated functional biomarkers of bladder cancers. Moreover, we are investigating the role of ChatGPT in omics research.
Research reported on this page was supported by National Institute of General Medical Sciences (NIGMS) of the National Institutes of Health under award number R15GM149587.
Latest News
-
September 14, 2021

New RIT visual modeling of coronavirus leads to discovery of behavior of second cellular ‘touchpoint’
WROC-TV features research by Gregory Babbitt, associate professor in the Thomas H. Gosnell School of Life Sciences; Patrick Rynkiewicz ’20 (bioinformatics and computational biology), ’21 MS (bioinformatics); Professor André Hudson, and Associate Professor Feng Cui.
-
August 17, 2021

RIT scientists model how coronavirus attaches itself to human cells
RIT scientists have uncovered new information about the way coronavirus and several of its variants attach to human cells. The researchers examined how coronaviruses use their spike proteins to attach themselves to the host cells they are attacking.
Sridevi Kayyur Subramanya '22 (bioinformatics MS)
Data Science Analyst at Navigate BioPharma Services, Inc.
Sheethal Umesh Nagalakshmi '21 (bioinformatics MS)
Computational Biologist at Beth Israel Deaconess Medical Center
Andrew Rosato '21 (bioinformatics MS)
Associate Bioinformatician at Massachusetts General Brigham
Peng Nien Yin '19 (bioinformatics MS)
Machine Learning Engineer at Paychex
Jimmy Zhang '18 (bioinformatics MS)
Senior Software Engineer at Athena Health
Feifei Bao '16 (bioinformatics MS)
Bioinformatician at Houston Methodist
Julia Freewoman '16 (bioinformatics MS)
Adjunct Lecturer at RIT
Peter LoVerso '15 (bioinformatics MS)
Bioinformatician at Precision Diagnostics
Gregory Wright '15 (bioinformatics MS)
Ph.D. student at University of Colorado
Bader A. Alharbi '14 (bioinformatics MS)
Ph.D. student at George Mason University






