Paul Craig Research Group
Contact
Paul Craig
Professor
585-475-6145
paul.craig@rit.edu
Developing interactive computer programs that simulate separations processes encountered in biochemistry and proteomics.
Overview
 My name is Dr. Paul A. Craig and I am a Professor of Biochemistry and Bioinformatics at the Rochester Institute of Technology located in Rochester, New York. I am also the Department Head for the School of Chemistry and Materials Science in the College of Science at RIT.
My name is Dr. Paul A. Craig and I am a Professor of Biochemistry and Bioinformatics at the Rochester Institute of Technology located in Rochester, New York. I am also the Department Head for the School of Chemistry and Materials Science in the College of Science at RIT.
My research involves developing interactive computer programs that simulate separations processes encountered in biochemistry and proteomics. To date we have developed simulations of ion exchange chromatography, one dimensional electrophoresis and two dimensional electrophoresis.
BASIL
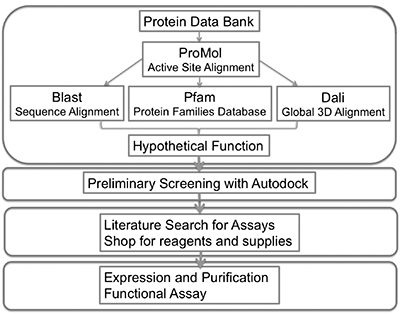 BASIL is an acronym for Biochemistry Authentic Scientific Inquiry Lab. In the BASIL curriculum, our aim is to get students to transition from thinking like students to thinking like scientists. Students will analyze proteins with known structures but unknown functions using computational analyses and wet-lab techniques. BASIL is designed for undergraduate biochemistry lab courses but can be adapted to first-year (or even high school) settings, as well as upper-level undergraduate or graduate coursework. It is targeted to students in biology, biochemistry, chemistry, or related majors.
BASIL is an acronym for Biochemistry Authentic Scientific Inquiry Lab. In the BASIL curriculum, our aim is to get students to transition from thinking like students to thinking like scientists. Students will analyze proteins with known structures but unknown functions using computational analyses and wet-lab techniques. BASIL is designed for undergraduate biochemistry lab courses but can be adapted to first-year (or even high school) settings, as well as upper-level undergraduate or graduate coursework. It is targeted to students in biology, biochemistry, chemistry, or related majors.
The curriculum is flexible and can be adapted to match the available facilities, the strengths of the instructor, and the learning goals of a course and institution. These lessons are often used as part of upper-level laboratory coursework with at least one semester of biochemistry as a pre-requisite or co-requisite. The lab has been designed for classes ranging from 10-24 students (working in teams of two or three) per lab section.
Python Scripting for BMB
 All scientists need to learn computer scripting/coding skills to remain competitive. Most undergraduate programs do not include coding skills in coursework for biology or chemistry majors, yet we hear of a need for basic coding skills from graduates who enter industry and those who go on to graduate school. During the spring of 2021, I worked with Jessica Nash at the Molecular Sciences Software Institute (MolSSI) to develop a Python scripting workshop. My goal is to introduce scientists from all professional levels (undergraduate, graduate, post-doc, faculty member, industrial scientist) to the use of Python programming in Jupyter notebooks, thereby enabling them to start taking advantage of computational power and flexibility that far exceeds data analysis and display tools found in Microsoft Excel and Apple Numbers.
All scientists need to learn computer scripting/coding skills to remain competitive. Most undergraduate programs do not include coding skills in coursework for biology or chemistry majors, yet we hear of a need for basic coding skills from graduates who enter industry and those who go on to graduate school. During the spring of 2021, I worked with Jessica Nash at the Molecular Sciences Software Institute (MolSSI) to develop a Python scripting workshop. My goal is to introduce scientists from all professional levels (undergraduate, graduate, post-doc, faculty member, industrial scientist) to the use of Python programming in Jupyter notebooks, thereby enabling them to start taking advantage of computational power and flexibility that far exceeds data analysis and display tools found in Microsoft Excel and Apple Numbers.
The workshop includes the following modules: Introduction, File Parsing, Processing Multiple Files and Writing Files, Working with Pandas, Linear Regression, Creating Plots in Jupyter Notebooks, and Nonlinear Regression.
- Github Repository for Python Scripting for Biochemistry & Molecular Biology
- Jupyter Book for Python Scripting for Biochemistry & Molecular Biology
- IQB Crash Course on Python Scripting for BMB
Electro2D-Tandem MS
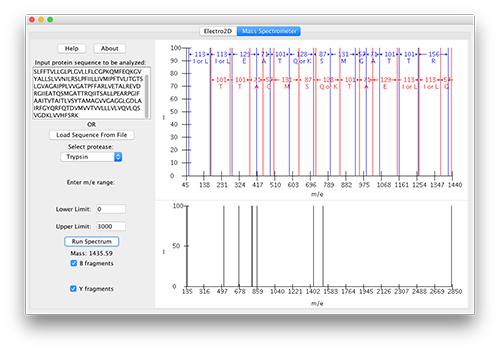 Electro2D-Tandem MS is a simulation of two-dimensional electrophoresis (2DE) - tandem mass spectroscopy (tandem MS), a tool that is used to separate and identify proteins from complex mixtures. This program was created entirely by students from programs in chemistry and computer science at RIT and is written in Java. Three notable contributors are Janine Garnham, who wrote the initial code for protein separation by 2DE; Jill Zapoticznyj, who started from Janine's code to build the graphical user interface for the simulation, and Amanda Fisher, who updated the code to make it work on any operating system and then integrated the protein separation by 2DE with Tandem Mass Spectrometry, which performs protein sequencing that can be used for protein identification and bioinformatics searching.
Electro2D-Tandem MS is a simulation of two-dimensional electrophoresis (2DE) - tandem mass spectroscopy (tandem MS), a tool that is used to separate and identify proteins from complex mixtures. This program was created entirely by students from programs in chemistry and computer science at RIT and is written in Java. Three notable contributors are Janine Garnham, who wrote the initial code for protein separation by 2DE; Jill Zapoticznyj, who started from Janine's code to build the graphical user interface for the simulation, and Amanda Fisher, who updated the code to make it work on any operating system and then integrated the protein separation by 2DE with Tandem Mass Spectrometry, which performs protein sequencing that can be used for protein identification and bioinformatics searching.
Human Visualization Project
A team of faculty and students from different colleges (College of Science, College of Art and Design, Golisano College of Computing and Information Sciences, Kate Gleason College of Engineering) and departments (Medical Illustration, Mechanical Engineering, Imaging Science, Biological Sciences, Biochemistry) at RIT participate in this project. Our goal is to create a 3D virtual human all the way from gross anatomy to organs, tissues, cells, and molecules.
JBioFramework
JBioFramework (JBF) is a collection of chemical separations simulations (Electrophoresis, Chromatography and Mass Spectrometry) that can enable students to experience the interface and results with these techniques in a safe, free environment. A full collection of the simulations can be downloaded from Sourceforge. It is written in the Java programming language and will run on computers with operating systems that have the Java Virtual Machine installed.




